Index
Welcome to the documentation for MAVEN.
Please use the navigation side-bar to access installation instructions, tutorials, and FAQs/Troubleshooting.
What is MAVEN?
MAVEN, or Mechanism of Action Visualisation and ENrichment, is an R Shiny app which is aimed at researchers wanting to understand the mechanism of action of a compound of interest. It requires 3 types of input from the user; a compound chemical structure, compound-induced gene expression data, and a prior knowledge network (we include the Omnipath [1] network with the package). MAVEN is tested and runs on Linux and Mac OS.
How does MAVEN work?
MAVEN was written to enable those without prior bioinformatics expertise to be able to use the different tools integrated within the software. The tools are DoRoThEA [2], PROGENy [3], CARNIVAL [4] and PIDGIN [5]. An overview of the tools and how they are integrated together in MAVEN is displayed below.
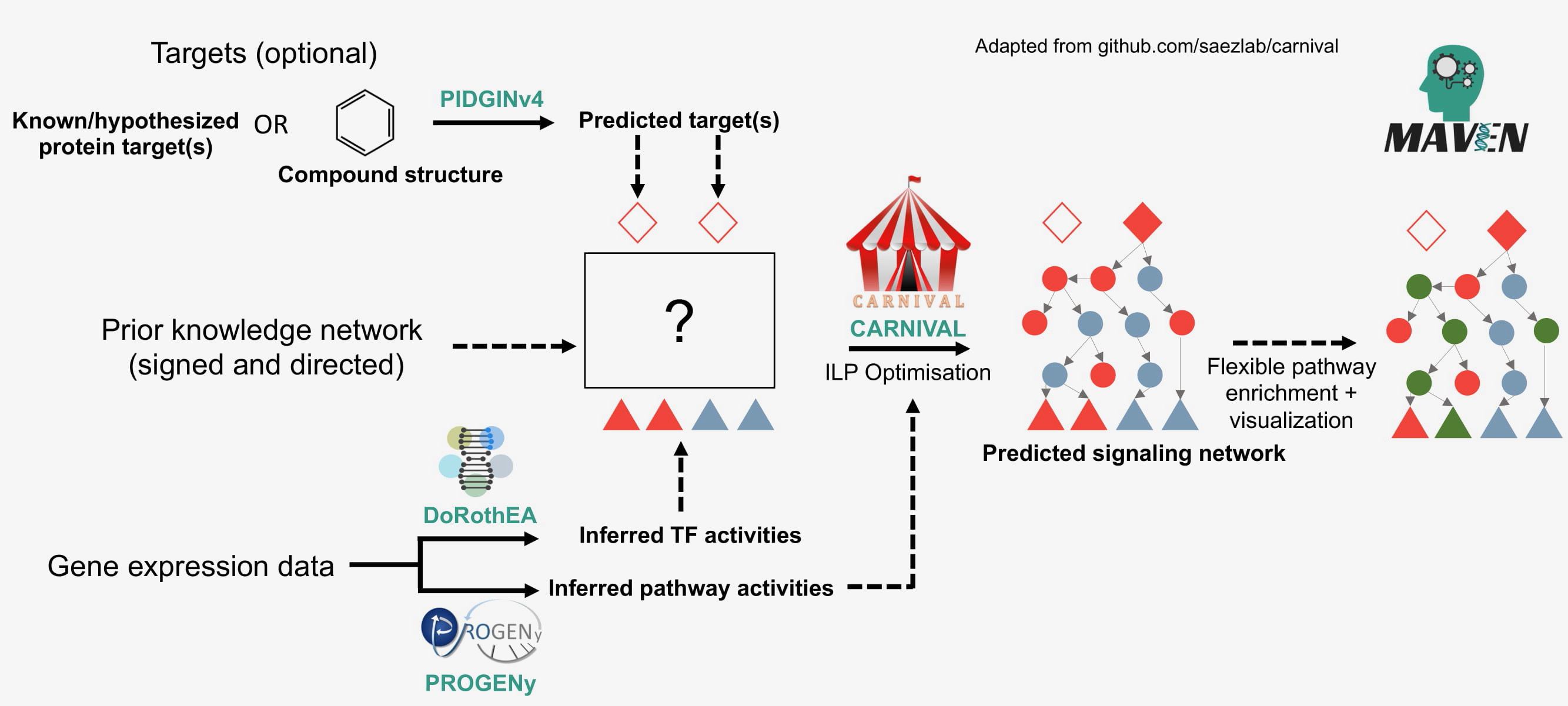
If you are unfamiliar with the tools used in MAVEN, we recommend you begin by following the tutorial.
What can I do with MAVEN?
MAVEN can be used to generate hypotheses for compound mechanism of action (or off-target effects/toxicity) which are both detailed and directly testable in the lab. Please see the Tutorial to learn more.
How can I cite MAVEN?
You can cite our pre-print if you use MAVEN in your work!
References
[1] D Turei, T Korcsmaros and J Saez-Rodriguez (2016). OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nature Methods 13 (12).
[2] L Garcia-Alonso, C. H. Holland, M. M. Ibrahim, D Turei and J Saez-Rodriguez (2019). Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res 29:1363-1375.
[3] M Schubert, B Klinger, M Klünemann, A Sieber, F Uhlitz, S Sauer, M. J. Garnett, N Blüthgen and J Saez-Rodriguez (2018). Perturbation-response genes reveal signaling footprints in cancer gene expression. Nature Comms 9 (20).
[4] A Liu, P Trairatphisan, E Gjerga, A Didangelos, J Barratt and J Saez-Rodriguez (2019). From expression footprints to causal pathways: contextualizing large signaling networks with CARNIVAL. npj Systems Biology and Applications 5 (40).
[5] L. H. Mervin, A. M. Afzal, G. Drakakis, R. Lewis, O. Engkvist and A. Bender (2015). Target prediction utilising negative bioactivity data covering large chemical space. Journal of Cheminformatics 7 (51).